|
||
|
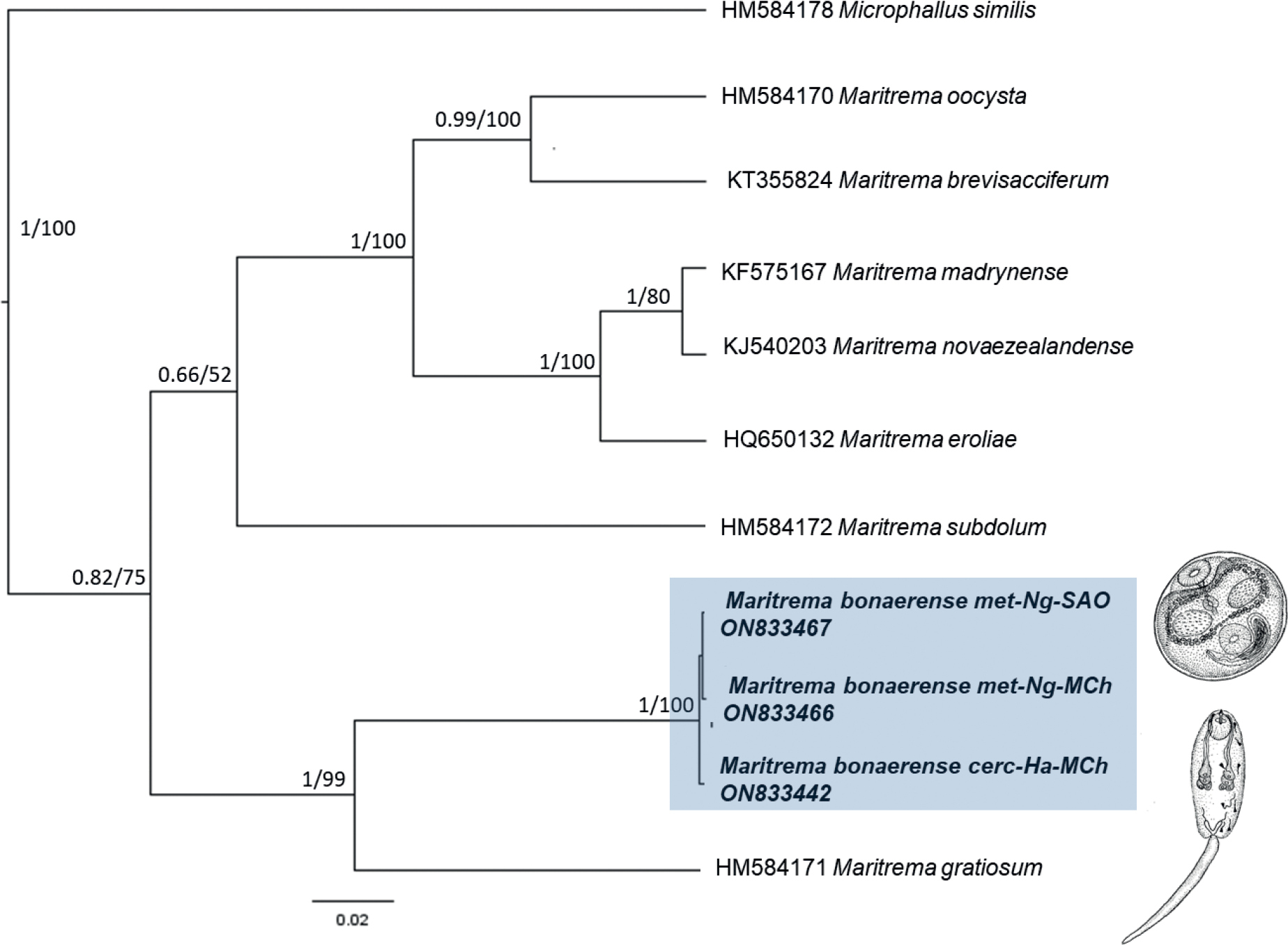
Phylogram for Maritrema species (Microphallus similis as outgroup), inferred by ML/BI of sequence data for ITS2 of the rRNA genes. The newly generated sequences are indicated in bold. Values on the branches correspond to posterior probabilities > 0.85 followed by bootstrap support > 60. Values below these thresholds were not reported. Abbreviations: cerc-cercaria, met-metacercaria, Ng-Neohelice granulata, Ha-Heleobia australis, MCh-Mar Chiquita Lagoon, SAO-San Antonio Oeste. Drafts of life stages extracted from Etchegoin and Martorelli (1997). |