|
||
|
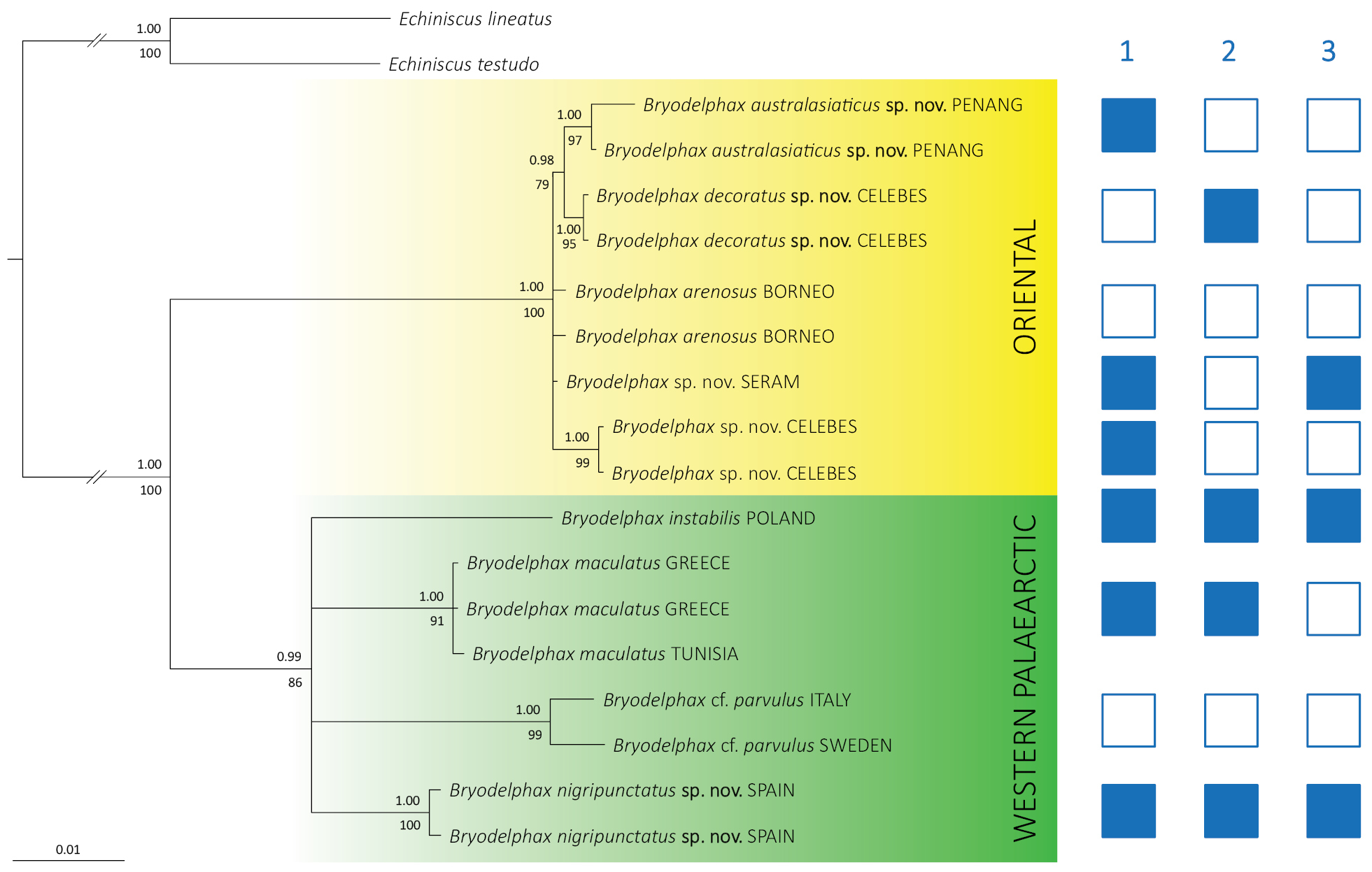
BI and ML concatenated (18S rRNA + 28S rRNA) phylogenetic tree of Bryodelphax Thulin, 1928; Echiniscus spp. were used as outgroup taxa. Bayesian posterior probability values (≥ 0.90) are given above tree branches, whereas ML bootstrap support values (≥ 70) are below branches. The scale bar represents 0.01 substitutions/site in the Bayesian tree. Trait mapping (blue square – presence, empty square – absence): 1 – ventral plates; 2 – granules on dorsal plates; 3 – males in population. |