|
||
|
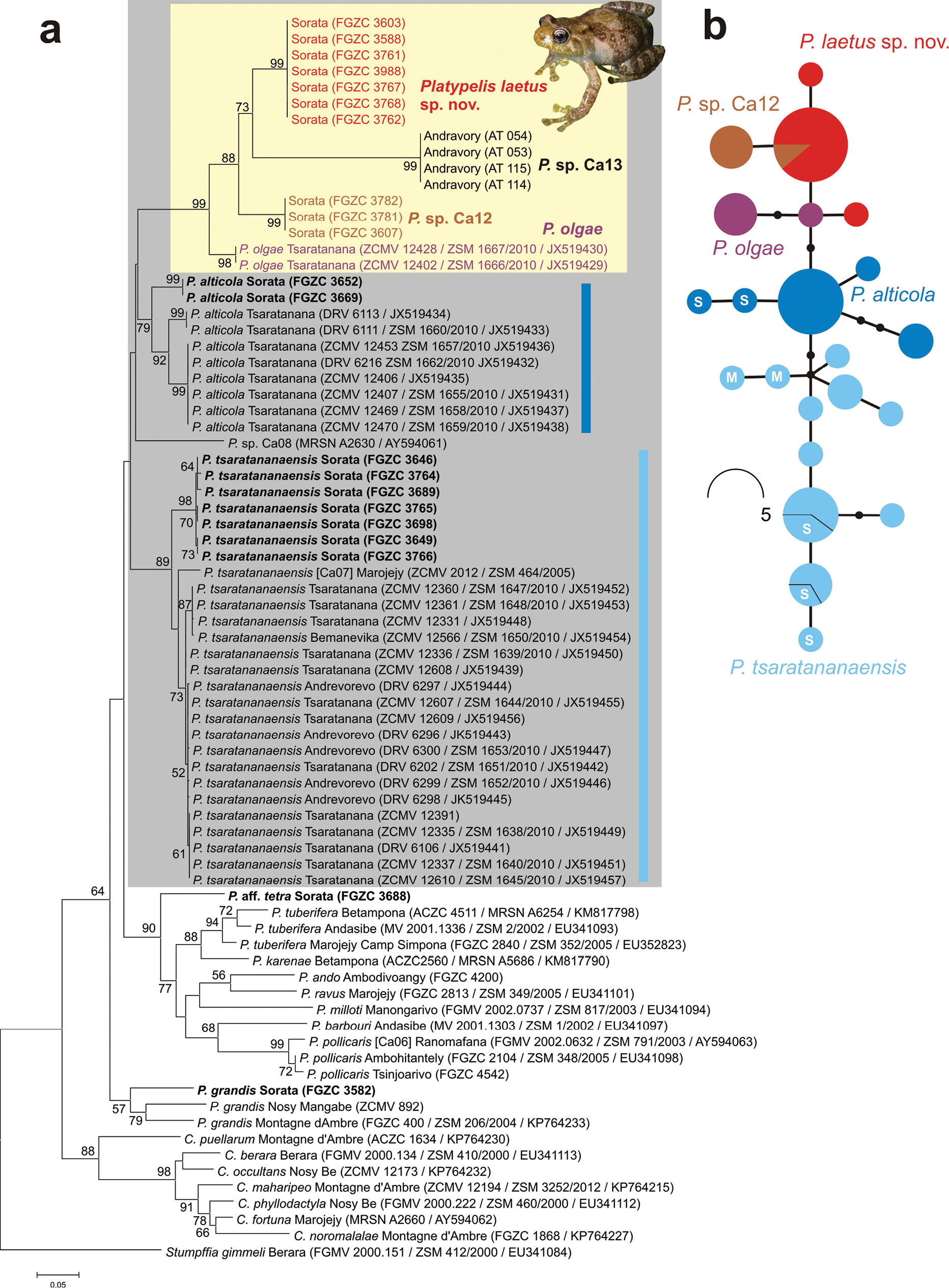
Molecular differentiation of species of Platypelis, with an emphasis on samples related to P. alticola, P. tsaratananaensis, and P. olgae (the highland cluster, in the gray box). a Maximum Likelihood tree calculated from a 658 bp alignment of the mitochondrial 16S rRNA gene (5ʹ segment of the gene). Numbers at nodes are support values from a bootstrap analysis (1000 replicates) in percent (not shown if below 50%). A sequence of Stumpffia gimmeli was included as the outgroup. For each sample, voucher number and if available, GenBank accession number is given. Samples outside the P. olgae clade (yellow box) that are highlighted in bold are newly sequenced specimens from the Sorata Massif. b Allele (haplotype) network based on 433 bp of the nuclear RAG-1 gene (phased sequences, hence each individual represented with two alleles in the network). In the P. alticola and P. tsaratananaensis clusters, an S marks alleles in samples from Sorata and an M marks samples from Marojejy. |