|
||
|
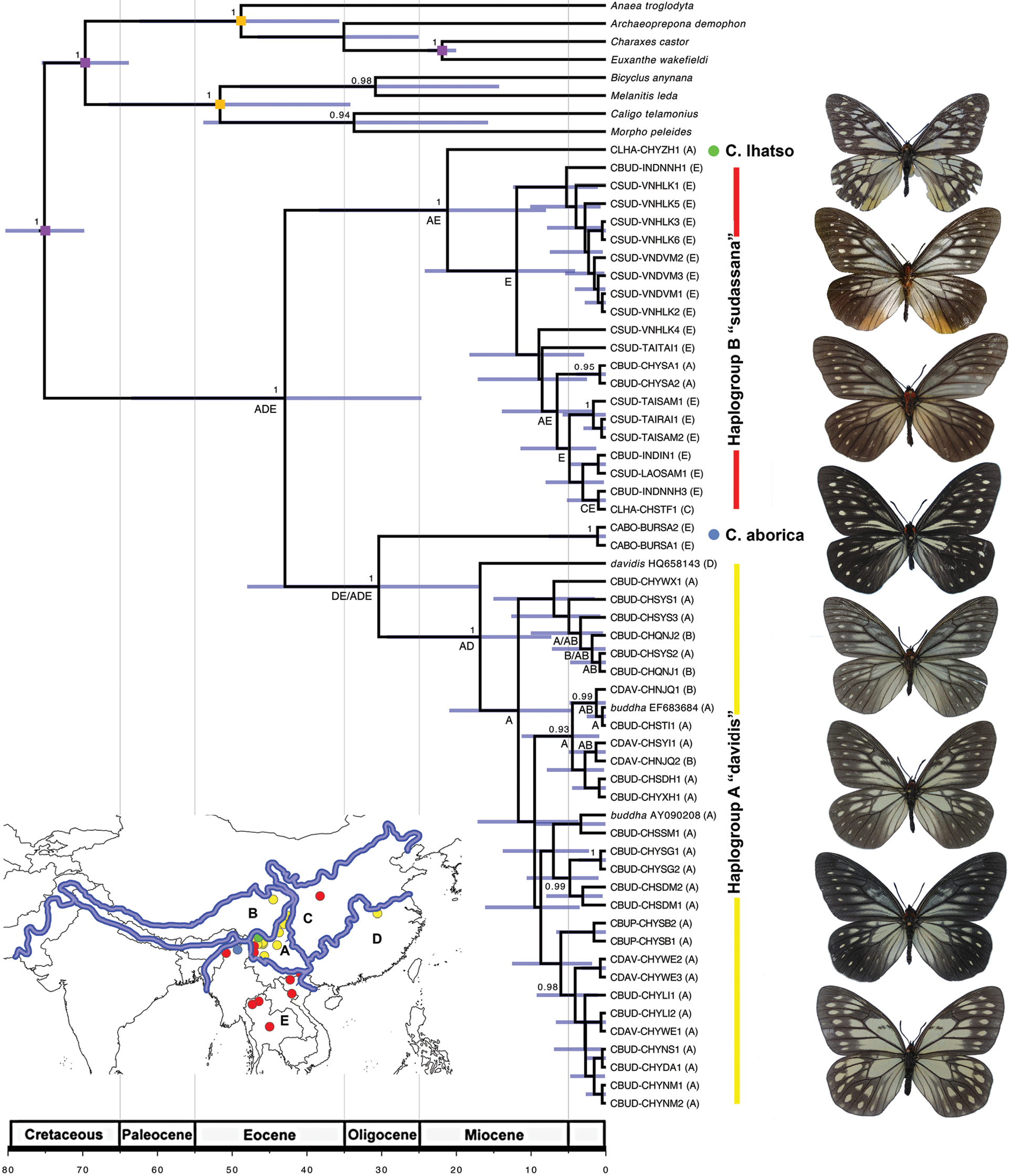
Bayesian phylogeny of Calinaga estimated in BEAST using concatenated data. Purple squares are calibration points (root: 75 ± 3; Satyrinae + Charaxinae 70 ± 3.5, Charaxes + Euxanthe 22 ± 1). Monophyly was enforced on nodes marked with orange squares. The inset map shows the biogeographic regions used in DIVA analysis: A) Southwestern China ecozone, B) Himalaya-Tibetan plateau region, C) Northern Sino-Himalaya, D) Southern Sino-Himalaya, E) Indochina. Colored dots correspond to haplogroups on the tree. |